Hit to Lead and Lead Optimization Services
After initial biological testing of selected compounds, the verified hits often undergo hit-to-lead analysis in order to progress several sets of compounds into just a few preclinical candidates.
Drug leads design
We design lead series for the target on the basis of verified hits, provided by a close collaboration between our partners and Aurora's chemistry teams, in two steps:
1. Creating new targeted libraries on the basis of hits verified in the initial biological testing stage.
2. Carrying out in silico screening of these compounds against the target.
ADME Modeling
Physiologically-based pharmacokinetic modeling (PBPK) is a mathematical modeling technique for prediction of the absorption, distribution, metabolism and excretion (ADME) of a compound in humans and other species used in pharmaceutical research.
ADME describes the disposition of a pharmaceutical compound within an organism. The four parameters influence the performance and pharmacological activity of the compound as a drug. PBPK or ADME modeling or ADME modeling or ADME prediction will be provided as shown below.
Our computer calculations we provided to estimate preclinical or non-clinical ADME parameters including:
- Absorption - maximum fraction absorbed, fraction absorbed, bioavailabilit;
- Distribution - organ/plasma partition coefficients and volume of distribution in steady state, concentration time curves in plasma and organs, active transport;
- Metabolism - predicting pharmacokinetic behavior on the basis of metabolism rates;
- Excretion - ways of excretion, enterohepatic cycle;
- Sensitivity analysis for interactions and individual variations;
- Pharmaceutical application - assessment of the best pharmaceutical form;
Quantitative Drug toxicity prediction (including LD50)
Our assay protocols have been developed to provide essential levels of information required for early assessment of compound toxicity and contain:
1. MRTD - maximum recommended therapeutic dose, MRDD - maximum recommended daily allowance, LD50.
2. To characterize the toxicity of a drug lead we screen the compound against a carefully selected panel of about 500 human proteins. The affinity constants (IC50, Kd, pKd) obtained in the calculations are then processed by our expert system and used to characterize the toxicity of the compound.
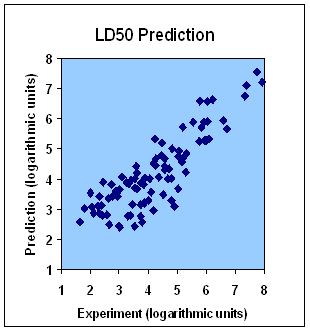
This service predicts drug toxicity and LD50. Toxicity prediction is provided both as an individual service and as an integral part of the compound profiling service. First, a toxicological profile is predicted by in silico screening of a drug candidate molecule against hundreds of diverse proteins representing different active site types of human proteome. The active site types that show high affinity for a compound (IC50) are considered as sensitive. In the second round of the toxicity assessment procedure all proteins similar to those with the sensitive active site types are included in screening. As a result, we obtain IC50 (IC50, represents the concentration of an inhibitor that is required for 50% inhibition of its target) of a tested compound for every protein from our toxicological protein platform. On the basis of this information we draw a conclusion about adverse reaction targets of a compound and its possible adverse effects. On the third step, our expert system calculates LD50 on the basis of predicted affinities (The figure shows calculated values of LD50 plotted against experimental ones for 100 drug molecules taken from Drug Bank).
In contrast to in vitro/vivo toxicity testing, in silico compound profiling does not require synthesis or purchasing chemicals. In addition, determination of chronic toxicity and carcinogenicity takes between several months and several years, while toxicity prediction in silico LD50 takes several hours or days. Finally, LD50 prediction saves the lives of animals.
Patentability Check
Our patentability check includes knowledge mining for IP score optimization (SciFinder, Beilstein/MDL, Integrity).